harold fellermann
Synthetic Biology
Simbiotics: a multi-scale integrative platform for 3D modeling of bacterial populations
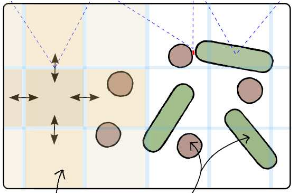
Simbiotics is a spatially explicit multi-scale modeling platform for the design, simulation and analysis of bacterial populations. Systems ranging from planktonic cells and colonies, to biofilm formation and development may be modeled. Representation of biological systems in Simbiotics is flexible, user-defined processes may be in a variety of forms depending on desired model abstraction. Simbiotics provides a library of modules such as cell geometries, physical force dynamics, genetic circuits, metabolic pathways, chemical diffusion and cell interactions. Model defined processes are integrated and scheduled for parallel multi-thead and multi-CPU execution. A virtual lab provides the modeler with analysis modules and some simulated lab equipment, enabling automation of sample interaction and data collection. An extendable and modular framework allows for the platform to be updated as novel models of bacteria are developed, coupled with an intuitive user interface to allow for model definitions with minimal programming experience. Simbiotics can integrate existing standards such as SBML, and process microscopy images to initialise the 3D spatial configuration of bacteria consortia. Two case studies, used to illustrate the platform flexibility, focus on the physical properties of the biosystems modeled. These pilot case studies demonstrate Simbiotics versatility in modeling and analysis of natural systems and as a CAD tool for synthetic biology.
J. Naylor, H. Fellermann, Y. Ding, W. K. Mohammed, N. S. Jakubovics, J. Mukherjee, C. A. Biggs, P. C. Wright, & N. Krasnogor, ACS Synthetic Biology, 2017
Designing uniquely addressable bio-orthogonal synthetic scaffolds for DNA and RNA origami
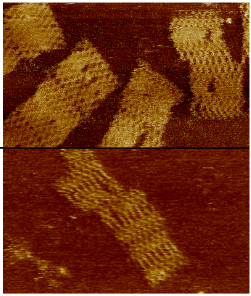
Nanotechnology and synthetic biology are rapidly converging, with DNA origami being one of the leading bridging technologies. DNA origami was shown to work well in a wide array of biotic environments. However, the large majority of extant DNA origami scaffolds utilize bacteriophages or plasmid sequences thus severely limiting its future applicability as bio-orthogonal nanotechnology platform. In this paper we present the design of biologically inert (i.e. "bio-orthogonal") origami scaffolds. The synthetic scaffolds have the additional advantage of being uniquely addressable (unlike biologically derived ones) and hence are better optimised for high-yield folding. We demonstrate our fully synthetic scaffold design with both DNA and RNA origamis and describe a protocol to produce these bio-orthogonal and uniquely addressable origami scaffolds.
J. Kozyra, A. Ceccarelli, E. Torelli, A. Lopiccolo, J. Gu, H. Fellermann, U. Stimming & N. Krasnogor, ACS Synthetic Biology, 2017
Toward programmable biology
This ACS Synthetic Biology special issue grew out of a focus workshop "Towards Programmable Biology" which was held in York, UK, on July 20, 2015 as part of the European Conference on Artificial Life ECAL 2015. The workshop gathered computer scientists, mathematicians, biologists, biophysicists, and the like to discuss routes towards general computability and programmability in biological substrates, and to demarcate key in vivo and in silico challenges of this novel research area. The issue also includes contributions from the 21st International Conference on DNA Computing and Molecular Programming DNA21, held on August 17-21, 2015 in Cambridge, MA, USA — a conference which revolves around similar questions.
H. Fellermann, O. Markovitch, C. Madsen, O. Gilfellon, A. Phillips, ACS Synthetic Biology, 2016
Listen to the interview in the ACS podcast
In vitro implementation of a stack data structure based on DNA strand displacement
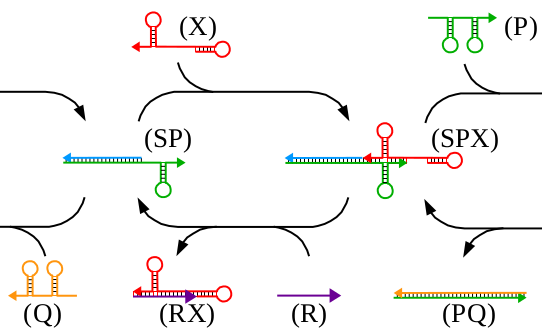
We present an implementation of an in vitro signal recorder based on DNA assembly and strand displacement. The signal recorder implements a stack data structure in which both data as well as operators are represented by single stranded DNA "bricks". The stack grows by adding push and write bricks and shrinks in last-in-first-out manner by adding pop and read bricks. We report the design of the signal recorder and its mode of operations and give experimental results from capillary electrophoresis as well as transmission electron microscopy that demonstrate the capability of the device to store and later release several successive signals. We conclude by discussing potential future improvements of our current results.
H. Fellermann, A. Lopiccolo, J. Kozyra & N. Krasnogor, Lect. Notes. Comp. Sci. 9726:87-98, 2016
Formalising Modularisation and Data Hiding for Synthetic Biology
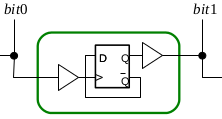
Biological systems employ compartmentalisation and other co-localisation strategies in order to orchestrate a multitude of biochemical processes by simultaneously enabling "data hiding" and modularisation. This paper presents recent research that embraces compartmentalisation and co-location as an organisational programmatic principle in synthetic biological and biomimetic systems. In these systems, artificial vesicles and synthetic minimal cells are envisioned as nanoscale reactors for programmable biochemical synthesis and as chassis for molecular information processing. We present P systems, brane calculi, and the recently developed chemtainer calculus as formal frameworks providing data hiding and modularisation and thus enabling the representation of highly complicated hierarchically organised compartmentalised reaction systems. We demonstrate how compartmentalisation can greatly reduce the complexity required to implement computational functionality, and how addressable compartments permits the scaling-up of programmable chemical synthesis.
H. Fellermann, M. Hadorn, R. Füchslin & N. Krasnogor, ACM J. Emerg. Tech. Comp. Sys 11(3):24, 2014
H. Fellermann & N. Krasnogor, Lect. Notes Comp. Sci. 8493:173-182, 2014
Modelling and stochastic simulation of synthetic biological Boolean gates
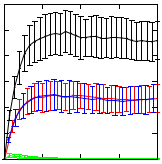
Synthetic Biology aspires to design and compose biological systems that implement specified behaviour in engineered biological system. When designing such systems, hypothesis testing via computational modelling and simulation is vital in order to reduce the need of costly wet lab experiments. As a case study, we here discuss the use of computational modelling and stochastic simulation of engineered genetic circuits that implement Boolean AND and OR gates that have been reported in the literature. We present performance analysis results for nine different state-of-the-art stochastic simulation algorithms and analyse the dynamic behaviour of the proposed gates. Stochastic simulations verify the desired functioning of the proposed gate designs.
D. Sanassy, H. Fellermann, N. Krasnogor, S. Konur, L. M. Mierla, M. Gheorghe, C. Ladroue & S. Kalvala, Proceedings of the 2014 IEEE International Conference on High Performance Computing and Communications pp. 404-408
Programming Chemistry in DNA Adressable Bioreactors
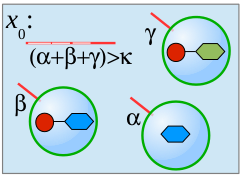
We present a formal calculus, termed chemtainer calculus, able to capture the complexity of compartmentalized reaction systems such as populations of possibly nested vesicular compartments. Compartments contain molecular cargo as well as surface markers in the form of DNA single strands. These markers serve as compartment addresses and allow for their targeted transport and fusion, thereby enabling reactions of previously separated chemicals. The overall system organization allows for the setup of programmable chemistry in microfluidic or other automated environments. We introduce a simple sequential programming language whose instructions are motivated by state-of-the-art microfluidic technology. Our approach integrates electronic control, chemical computing, and material production in a unified formal framework that is able to mimick the integrated computational and constructive capabilities of the subcellular matrix. We provide a non-deterministic semantics of our programming language that enables us to analytically derive the computational and constructive power of our machinary. This semantics is used to derive the sets of all constructable chemicals and supermolecular structures that emerge from different underlying instruction sets. Since our proofs are constructive, they can be utilized to automatically infer control programs for the construction of target structures from a limited set of resource molecules. Finally, we present an example of our framework from the area of oligosaccharide synthesis.
H. Fellermann & L. Cardelli, R. Soc. Interface 11(99), 2014
Specific and Reversible DNA-directed Self-Assembly of Oil-in-Water Emulsion Droplets
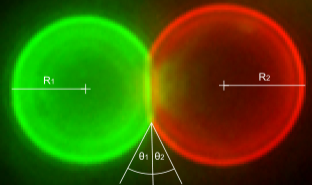
Higher-order structures that originate from the specific and reversible DNA-directed self-assembly of microscopic building blocks hold great promise for future technologies. Here, we functionalized biotinylated soft colloid oil-in-water emulsion droplets with biotinylated single-stranded DNA oligonucleotides using streptavidin as an intermediary linker. We show the components of this modular linking system to be stable and to induce sequence-specific aggregation of binary mixtures of emulsion droplets. Three length scales were thereby involved: nanoscale DNA base pairing linking microscopic building blocks resulted in macroscopic aggregates visible to the naked eye. The aggregation process was reversible by changing the temperature and electrolyte concentration and by the addition of competing oligonucleotides. The system was reset and reused by subsequent refunctionalization of the emulsion droplets. DNA-directed self-assembly of oil-in-water emulsion droplets, therefore, offers a solid basis for programmable and recyclable soft materials that undergo structural rearrangements on demand and that range in application from information technology to medicine.
M. Hadorn, E. Bönzli, K. T. Sørensen, H. Fellermann, P. Eggenberger-Hotz & M. Hanczyc, Proc. Nat. Acad. Sci. USA 109(47), 2012
