harold fellermann
Origins of Life and Molecular Replicators
Population Dynamics of Autocatalytic Sets in a Compartmentalized Spatial World
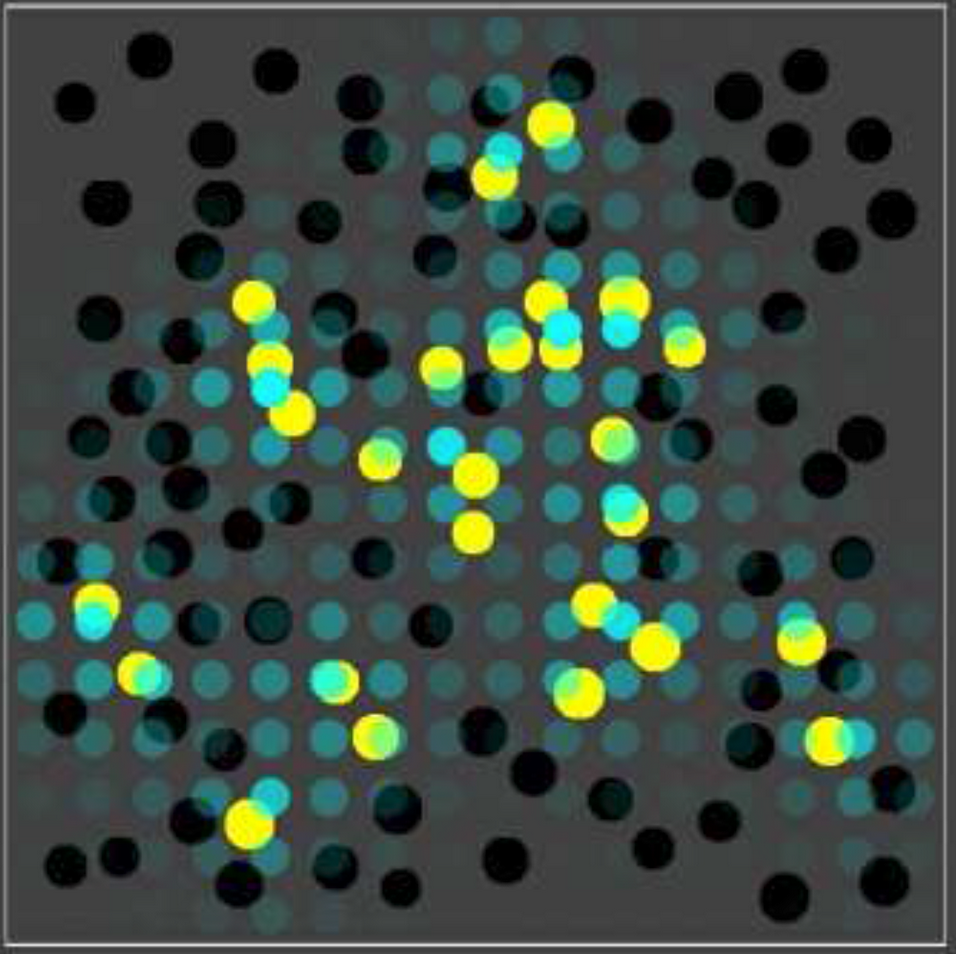
Autocatalytic sets are self-sustaining and collectively catalytic chemical reaction networks which are believed to have played an important role in the origin of life. Simulation studies have shown that autocatalytic sets are, in principle, evolvable if multiple autocatalytic subsets can exist in different combinations within compartments, i.e., so-called protocells. However, these previous studies have so far not explicitly modeled the emergence and dynamics of autocatalytic sets in populations of compartments in a spatial environment. Here, we use a recently developed software tool to simulate exactly this scenario, as an important first step towards more realistic simulations and experiments on autocatalytic sets in protocells.
W. Hordijk, J. Naylor, N. Krasnogor & H. Fellermann Life 8(3), 33, 2018
Sequence selection by dynamical symmetry breaking in an autocatalytic binary polymer model
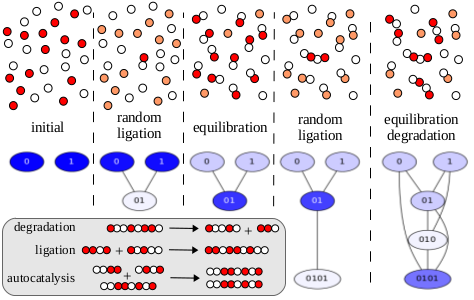
Template-directed replication of nucleic acids is at the essence of all living beings and a major milestone for any origin of life scenario. We present an idealized model of prebiotic sequence replication, where binary polymers act as templates for their autocatalytic replication, thereby serving as each others reactants and products in an intertwined molecular ecology. Our model demonstrates how autocatalysis alters the qualitative and quantitative system dynamics in counterintuitive ways. Most notably, numerical simulations reveal a very strong intrinsic selection mechanism that favors the appearance of a few population structures with highly ordered and repetitive sequence patterns when starting from a pool of monomers. We demonstrate both analytically and through simulation how this "selection of the dullest" is caused by continued symmetry breaking through random fluctuations in the transient dynamics that are amplified by autocatalysis and eventually propagate to the population level. The impact of these observations on related prebiotic mathematical models is discussed.
H. Fellermann, S. Tanaka & S. Rasmussen, Physical Review E 96, 062407, 2017
Sequence selection in an autocatalytic binary polymer model
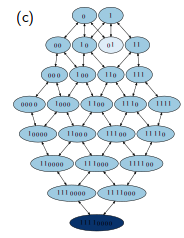
An autocatalytic pattern matching polymer system is studied as an abstract model for chemical ecosystem evolution. Highly ordered populations with particular sequence patterns appear spontaneously out of a vast number of possible states. The interplay between the selected microscopic sequence patterns and the macroscopic cooperative structures is examined. Stability, fluctuations, and evolutionary selection mechanisms are investigated for the involved self-organizing processes.
S. Tanaka, H. Fellermann & S. Rasmussen, Europhys. Lett. 107(2), 28004, 2014
On the Growth Rate of Non-Enzymatic Molecular Replicators
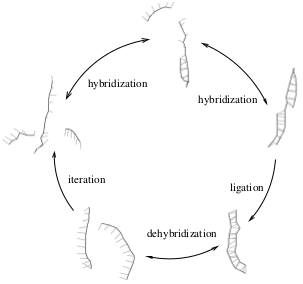
It is well known that non-enzymatic template directed molecular replicators X + nO -> 2X exhibit parabolic growth d[X]/dt -> k[X]1/2. Here, we analyze the dependence of the effective replication rate constant k on hybridization energies, temperature, strand length, and sequence composition. First we derive analytical criteria for the replication rate k based on simple thermodynamic arguments. Second we present a Brownian dynamics model for oligonucleotides that allows us to simulate their diffusion and hybridization behavior. The simulation is used to generate and analyze the effect of strand length, temperature, and to some extent sequence composition, on the hybridization rates and the resulting optimal overall rate constant k. Combining the two approaches allows us to semi-analytically depict a replication rate landscape for template directed replicators. The results indicate a clear replication advantage for longer strands at lower temperatures in the regime where the ligation rate is rate limiting. Further the results indicate the existence of an optimal replication rate at the boundary between the two regimes where the ligation rate and the dehybridization rates are rate limiting.
H. Fellermann & S. Rasmussen, Entropy 13(10):1882-1903, 2011
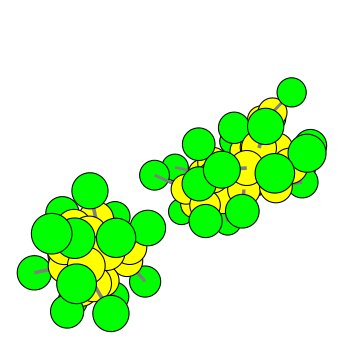
Life Cycle of a Minimal Protocell – A Dissipative Particle Dynamics Study
Cross-reactions and other systematic difficulties generated by the coupling of functional chemical subsystems pose the largest challenge for assembling a viable protocell in the laboratory. Our current work seeks to identify and clarify such key issues as we represent and analyze in simulation a full implementation of a minimal protocell. Using a 3D dissipative particle dynamics simulation method, we are able to address the coupled diffusion, self-assembly, and chemical reaction processes required to model a full life cycle of a protocell composed of coupled genetic, metabolic, and container subsystems. Utilizing this minimal structural and functional representation of the constituent molecules, their interactions, and their reactions, we identify and explore the nature of the many linked processes for the full protocellular system. Obviously the simplicity of this simulation method combined with the inherent system complexity prevents us from expecting quantitative simulation predictions from these investigations. However, we report important findings on systemic processes, some previously predicted and some newly discovered, as we couple the protocellular self-assembly processes and chemical reactions.
H. Fellermann, S. Rasmussen, H.-J. Ziock & R. Solé, Artif. Life 13(4):319-345, 2007
Minimal model of self-replicating nanocells – A physically embodied, information-free scenario
The building of minimal self-reproducing systems with a physical embodiment (generically called protocells) is a great challenge, with implications for both theory and applied sciences. Although the classical view of a living protocell assumes that it includes information-carrying molecules as an essential ingredient, a dividing cell-like structure can be built from a metabolism-container coupled system only. An example of such a system, modelled with dissipative particle dynamics, is presented here. This article demonstrates how a simple coupling between a precursor molecule and surfactant molecules forming micelles can experience a growth-division cycle in a predictable manner, and analyses the influence of crucial parameters on this replication cycle. Implications of these results for origins of cellular life and living technology are outlined.
H. Fellermann & R. Solé, Philos. Trans. R. Soc. Ser. B 362(1486):1803-1811, 2007
