harold fellermann
Research Interests
Driving force behind my endeavors in science is the desire to better understand the creative potential of Nature.
Population Dynamics of Autocatalytic Sets in a Compartmentalized Spatial World
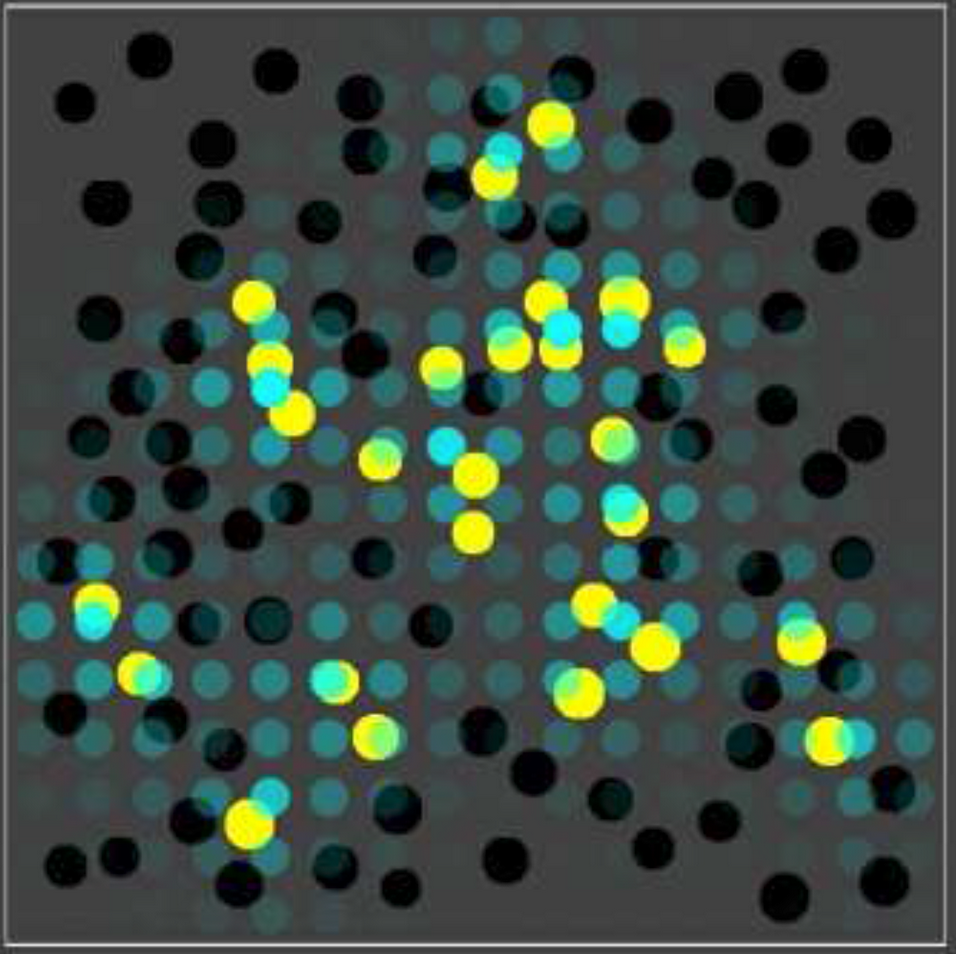
Autocatalytic sets are self-sustaining and collectively catalytic chemical reaction networks which are believed to have played an important role in the origin of life. Simulation studies have shown that autocatalytic sets are, in principle, evolvable if multiple autocatalytic subsets can exist in different combinations within compartments, i.e., so-called protocells. However, these previous studies have so far not explicitly modeled the emergence and dynamics of autocatalytic sets in populations of compartments in a spatial environment. Here, we use a recently developed software tool to simulate exactly this scenario, as an important first step towards more realistic simulations and experiments on autocatalytic sets in protocells.
W. Hordijk, J. Naylor, N. Krasnogor & H. Fellermann Life 8(3), 33, 2018
Sequence selection by dynamical symmetry breaking in an autocatalytic binary polymer model
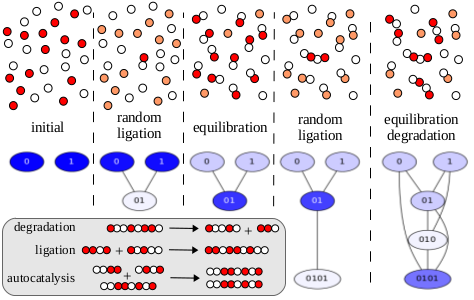
Template-directed replication of nucleic acids is at the essence of all living beings and a major milestone for any origin of life scenario. We present an idealized model of prebiotic sequence replication, where binary polymers act as templates for their autocatalytic replication, thereby serving as each others reactants and products in an intertwined molecular ecology. Our model demonstrates how autocatalysis alters the qualitative and quantitative system dynamics in counterintuitive ways. Most notably, numerical simulations reveal a very strong intrinsic selection mechanism that favors the appearance of a few population structures with highly ordered and repetitive sequence patterns when starting from a pool of monomers. We demonstrate both analytically and through simulation how this "selection of the dullest" is caused by continued symmetry breaking through random fluctuations in the transient dynamics that are amplified by autocatalysis and eventually propagate to the population level. The impact of these observations on related prebiotic mathematical models is discussed.
H. Fellermann, S. Tanaka & S. Rasmussen, Physical Review E 96, 062407, 2017
Simbiotics: a multi-scale integrative platform for 3D modeling of bacterial populations
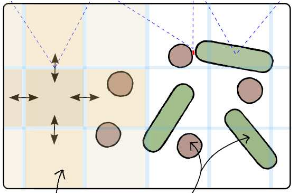
Simbiotics is a spatially explicit multi-scale modeling platform for the design, simulation and analysis of bacterial populations. Systems ranging from planktonic cells and colonies, to biofilm formation and development may be modeled. Representation of biological systems in Simbiotics is flexible, user-defined processes may be in a variety of forms depending on desired model abstraction. Simbiotics provides a library of modules such as cell geometries, physical force dynamics, genetic circuits, metabolic pathways, chemical diffusion and cell interactions. Model defined processes are integrated and scheduled for parallel multi-thead and multi-CPU execution. A virtual lab provides the modeler with analysis modules and some simulated lab equipment, enabling automation of sample interaction and data collection. An extendable and modular framework allows for the platform to be updated as novel models of bacteria are developed, coupled with an intuitive user interface to allow for model definitions with minimal programming experience. Simbiotics can integrate existing standards such as SBML, and process microscopy images to initialise the 3D spatial configuration of bacteria consortia. Two case studies, used to illustrate the platform flexibility, focus on the physical properties of the biosystems modeled. These pilot case studies demonstrate Simbiotics versatility in modeling and analysis of natural systems and as a CAD tool for synthetic biology.
J. Naylor, H. Fellermann, Y. Ding, W. K. Mohammed, N. S. Jakubovics, J. Mukherjee, C. A. Biggs, P. C. Wright, & N. Krasnogor, ACS Synthetic Biology, 2017
Optimizing nucleic acid sequences for a molecular data recorder
We recently reported the design for a DNA nano-device that can record and store molecular signals. Here we present an evolutionary algorithm tailored to optimising nucleic acid sequences that predictively fold into our desired target structures. In our approach, a DNA device is first specified abstractly: the topology of the individual strands and their desired foldings into multi-strand complexes are described at the domain-level. Initially, this design is decomposed into a set of pairwise strand interactions. Then, we optimize candidate domains, such that the resulting sequences fold with high accuracy into desired target structures both (a) individually and (b) jointly, but also (c) to show high affinity for binding desired partners and simultaneously low affinity to bind with any undesired partner. As optimization heuristic we use a genetic algorithm that employs a linear combination of the above scores. Our algorithm was able to generate DNA sequences that satisfy all given criteria. Even though we cannot establish the theoretically achievable optima (as this would require exhaustive search), our solutions score 90% of an upper bound that ignores conflicting objectives. We envision that this approach can be generalized towards a broad class of toehold-mediated strand displacement systems.
J. Kozyra, H. Fellermann, B. Shirt-Ediss, A. Lopiccolo & N. Krasnogor, Proceedings of the Genetic and Evolutionary Computation Conference GECCO, 2017
Designing uniquely addressable bio-orthogonal synthetic scaffolds for DNA and RNA origami
Nanotechnology and synthetic biology are rapidly converging, with DNA origami being one of the leading bridging technologies. DNA origami was shown to work well in a wide array of biotic environments. However, the large majority of extant DNA origami scaffolds utilize bacteriophages or plasmid sequences thus severely limiting its future applicability as bio-orthogonal nanotechnology platform. In this paper we present the design of biologically inert (i.e. "bio-orthogonal") origami scaffolds. The synthetic scaffolds have the additional advantage of being uniquely addressable (unlike biologically derived ones) and hence are better optimised for high-yield folding. We demonstrate our fully synthetic scaffold design with both DNA and RNA origamis and describe a protocol to produce these bio-orthogonal and uniquely addressable origami scaffolds.
J. Kozyra, A. Ceccarelli, E. Torelli, A. Lopiccolo, J. Gu, H. Fellermann, U. Stimming & N. Krasnogor, ACS Synthetic Biology, 2017
Toward programmable biology
This ACS Synthetic Biology special issue grew out of a focus workshop "Towards Programmable Biology" which was held in York, UK, on July 20, 2015 as part of the European Conference on Artificial Life ECAL 2015. The workshop gathered computer scientists, mathematicians, biologists, biophysicists, and the like to discuss routes towards general computability and programmability in biological substrates, and to demarcate key in vivo and in silico challenges of this novel research area. The issue also includes contributions from the 21st International Conference on DNA Computing and Molecular Programming DNA21, held on August 17-21, 2015 in Cambridge, MA, USA — a conference which revolves around similar questions.
H. Fellermann, O. Markovitch, C. Madsen, O. Gilfellon, A. Phillips, ACS Synthetic Biology, 2016
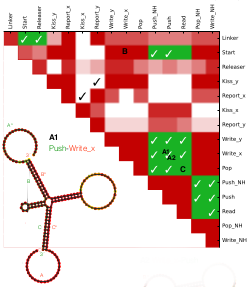
In vitro implementation of a stack data structure based on DNA strand displacement
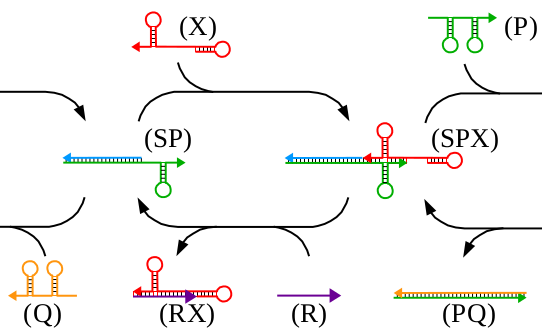
We present an implementation of an in vitro signal recorder based on DNA assembly and strand displacement. The signal recorder implements a stack data structure in which both data as well as operators are represented by single stranded DNA "bricks". The stack grows by adding push and write bricks and shrinks in last-in-first-out manner by adding pop and read bricks. We report the design of the signal recorder and its mode of operations and give experimental results from capillary electrophoresis as well as transmission electron microscopy that demonstrate the capability of the device to store and later release several successive signals. We conclude by discussing potential future improvements of our current results.
H. Fellermann, A. Lopiccolo, J. Kozyra & N. Krasnogor, Lect. Notes. Comp. Sci. 9726:87-98, 2016
Formalising Modularisation and Data Hiding for Synthetic Biology
Biological systems employ compartmentalisation and other co-localisation strategies in order to orchestrate a multitude of biochemical processes by simultaneously enabling "data hiding" and modularisation. This paper presents recent research that embraces compartmentalisation and co-location as an organisational programmatic principle in synthetic biological and biomimetic systems. In these systems, artificial vesicles and synthetic minimal cells are envisioned as nanoscale reactors for programmable biochemical synthesis and as chassis for molecular information processing. We present P systems, brane calculi, and the recently developed chemtainer calculus as formal frameworks providing data hiding and modularisation and thus enabling the representation of highly complicated hierarchically organised compartmentalised reaction systems. We demonstrate how compartmentalisation can greatly reduce the complexity required to implement computational functionality, and how addressable compartments permits the scaling-up of programmable chemical synthesis.
H. Fellermann, M. Hadorn, R. Füchslin & N. Krasnogor, ACM J. Emerg. Tech. Comp. Sys 11(3):24, 2014
H. Fellermann & N. Krasnogor, Lect. Notes Comp. Sci. 8493:173-182, 2014
Modelling and stochastic simulation of synthetic biological Boolean gates
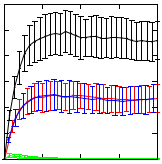
Synthetic Biology aspires to design and compose biological systems that implement specified behaviour in engineered biological system. When designing such systems, hypothesis testing via computational modelling and simulation is vital in order to reduce the need of costly wet lab experiments. As a case study, we here discuss the use of computational modelling and stochastic simulation of engineered genetic circuits that implement Boolean AND and OR gates that have been reported in the literature. We present performance analysis results for nine different state-of-the-art stochastic simulation algorithms and analyse the dynamic behaviour of the proposed gates. Stochastic simulations verify the desired functioning of the proposed gate designs.
D. Sanassy, H. Fellermann, N. Krasnogor, S. Konur, L. M. Mierla, M. Gheorghe, C. Ladroue & S. Kalvala, Proceedings of the 2014 IEEE International Conference on High Performance Computing and Communications pp. 404-408
Programming Chemistry in DNA Adressable Bioreactors
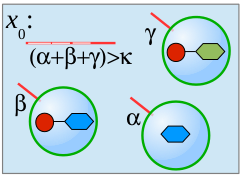
We present a formal calculus, termed chemtainer calculus, able to capture the complexity of compartmentalized reaction systems such as populations of possibly nested vesicular compartments. Compartments contain molecular cargo as well as surface markers in the form of DNA single strands. These markers serve as compartment addresses and allow for their targeted transport and fusion, thereby enabling reactions of previously separated chemicals. The overall system organization allows for the setup of programmable chemistry in microfluidic or other automated environments. We introduce a simple sequential programming language whose instructions are motivated by state-of-the-art microfluidic technology. Our approach integrates electronic control, chemical computing, and material production in a unified formal framework that is able to mimick the integrated computational and constructive capabilities of the subcellular matrix. We provide a non-deterministic semantics of our programming language that enables us to analytically derive the computational and constructive power of our machinary. This semantics is used to derive the sets of all constructable chemicals and supermolecular structures that emerge from different underlying instruction sets. Since our proofs are constructive, they can be utilized to automatically infer control programs for the construction of target structures from a limited set of resource molecules. Finally, we present an example of our framework from the area of oligosaccharide synthesis.
H. Fellermann & L. Cardelli, R. Soc. Interface 11(99), 2014
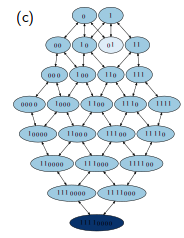
Sequence selection in an autocatalytic binary polymer model
An autocatalytic pattern matching polymer system is studied as an abstract model for chemical ecosystem evolution. Highly ordered populations with particular sequence patterns appear spontaneously out of a vast number of possible states. The interplay between the selected microscopic sequence patterns and the macroscopic cooperative structures is examined. Stability, fluctuations, and evolutionary selection mechanisms are investigated for the involved self-organizing processes.
S. Tanaka, H. Fellermann & S. Rasmussen, Europhys. Lett. 107(2), 28004, 2014
The MATCHIT Automaton: Exploiting Compartmentalization for the Synthesis of Branched Polymers
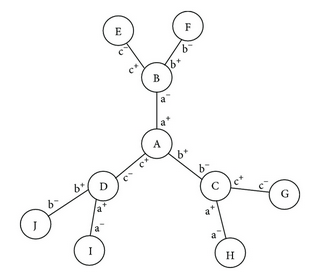
We propose an automaton, a theoretical framework that demonstrates how to improve the yield of the synthesis of branched chemical polymer reactions. This is achieved by separating substeps of the path of synthesis into compartments. We use chemical containers (chemtainers) to carry the substances through a sequence of fixed successive compartments. We describe the automaton in mathematical terms and show how it can be configured automatically in order to synthesize a given branched polymer target. The algorithm we present finds an optimal path of synthesis in linear time. We discuss how the automaton models compartmentalized structures found in cells, such as the endoplasmic reticulum and the Golgi apparatus, and we show how this compartmentalization can be exploited for the synthesis of branched polymers such as oligosaccharides. Lastly, we show examples of artificial branched polymers and discuss how the automaton can be configured to synthesize them with maximal yield.
M. Weyland, H. Fellermann, M. Hadorn, D. Sorek, D. Lancet, S. Rasmussen, & R. M. Füchslin, Comput. Math. Met. Med. 467428, 2013
Specific and Reversible DNA-directed Self-Assembly of Oil-in-Water Emulsion Droplets
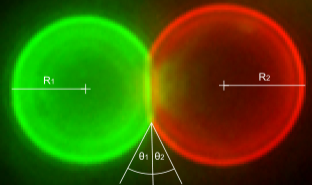
Higher-order structures that originate from the specific and reversible DNA-directed self-assembly of microscopic building blocks hold great promise for future technologies. Here, we functionalized biotinylated soft colloid oil-in-water emulsion droplets with biotinylated single-stranded DNA oligonucleotides using streptavidin as an intermediary linker. We show the components of this modular linking system to be stable and to induce sequence-specific aggregation of binary mixtures of emulsion droplets. Three length scales were thereby involved: nanoscale DNA base pairing linking microscopic building blocks resulted in macroscopic aggregates visible to the naked eye. The aggregation process was reversible by changing the temperature and electrolyte concentration and by the addition of competing oligonucleotides. The system was reset and reused by subsequent refunctionalization of the emulsion droplets. DNA-directed self-assembly of oil-in-water emulsion droplets, therefore, offers a solid basis for programmable and recyclable soft materials that undergo structural rearrangements on demand and that range in application from information technology to medicine.
M. Hadorn, E. Bönzli, K. T. Sørensen, H. Fellermann, P. Eggenberger-Hotz & M. Hanczyc, Proc. Nat. Acad. Sci. USA 109(47), 2012
DNA Self-Assembly and Computation Studied with a Coarse-grained Dynamic Bonded Model
We study DNA self-assembly and DNA computation using a coarse-grained DNA model within the directional dynamic bonding framework [C. Svaneborg, Comp. Phys. Comm. 183, 1793 (2012)]. In our model, a single nucleotide or domain is represented by a single interaction site. Complementary sites can reversibly hybridize and dehybridize during a simulation. This bond dynamics induces a dynamics of the angular and dihedral bonds, that model the collective effects of chemical structure on the hybridization dynamics. We use the DNA model to perform simulations of the self-assembly kinetics of DNA tetrahedra, an icosahedron, as well as strand displacement operations used in DNA computation.
C. Svaneborg, H. Fellermann & S. Rasmussen, Lect. Notes in Comput. Sc. 7433, 123, 2012
On the Growth Rate of Non-Enzymatic Molecular Replicators
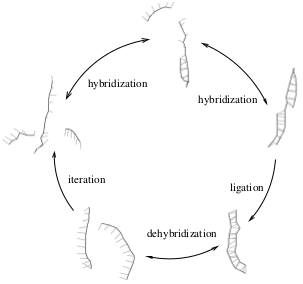
It is well known that non-enzymatic template directed molecular replicators X + nO -> 2X exhibit parabolic growth d[X]/dt -> k[X]1/2. Here, we analyze the dependence of the effective replication rate constant k on hybridization energies, temperature, strand length, and sequence composition. First we derive analytical criteria for the replication rate k based on simple thermodynamic arguments. Second we present a Brownian dynamics model for oligonucleotides that allows us to simulate their diffusion and hybridization behavior. The simulation is used to generate and analyze the effect of strand length, temperature, and to some extent sequence composition, on the hybridization rates and the resulting optimal overall rate constant k. Combining the two approaches allows us to semi-analytically depict a replication rate landscape for template directed replicators. The results indicate a clear replication advantage for longer strands at lower temperatures in the regime where the ligation rate is rate limiting. Further the results indicate the existence of an optimal replication rate at the boundary between the two regimes where the ligation rate and the dehybridization rates are rate limiting.
H. Fellermann & S. Rasmussen, Entropy 13(10):1882-1903, 2011
Coarse graining and scaling in dissipative particle dynamics
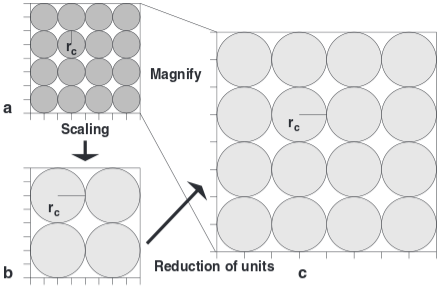
Dissipative particle dynamics (DPD) is now a well-established method for simulating soft matter systems. However, its applicability was recently questioned because some investigations showed an upper coarse-graining limit that would prevent the applicability of the method to the whole mesoscopic range. This article aims to re-establish DPD as a truly mesoscopic method by analyzing the problems reported by other authors and by presenting a scaling scheme that allows one to apply DPD simulations directly to any desired length scale.
R. Füchslin, H. Fellermann, A. Eriksson & H.-J. Ziock, J. Chem. Phys. 130(21):214102, 2009
Life Cycle of a Minimal Protocell – A Dissipative Particle Dynamics Study
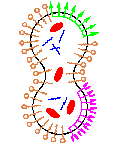
Cross-reactions and other systematic difficulties generated by the coupling of functional chemical subsystems pose the largest challenge for assembling a viable protocell in the laboratory. Our current work seeks to identify and clarify such key issues as we represent and analyze in simulation a full implementation of a minimal protocell. Using a 3D dissipative particle dynamics simulation method, we are able to address the coupled diffusion, self-assembly, and chemical reaction processes required to model a full life cycle of a protocell composed of coupled genetic, metabolic, and container subsystems. Utilizing this minimal structural and functional representation of the constituent molecules, their interactions, and their reactions, we identify and explore the nature of the many linked processes for the full protocellular system. Obviously the simplicity of this simulation method combined with the inherent system complexity prevents us from expecting quantitative simulation predictions from these investigations. However, we report important findings on systemic processes, some previously predicted and some newly discovered, as we couple the protocellular self-assembly processes and chemical reactions.
H. Fellermann, S. Rasmussen, H.-J. Ziock & R. Solé, Artif. Life 13(4):319-345, 2007
Minimal model of self-replicating nanocells – A physically embodied, information-free scenario
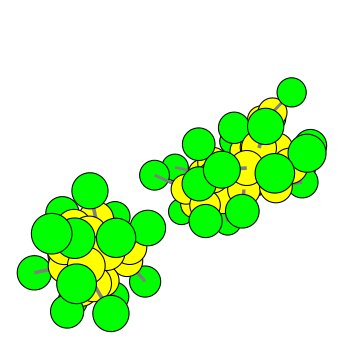
The building of minimal self-reproducing systems with a physical embodiment (generically called protocells) is a great challenge, with implications for both theory and applied sciences. Although the classical view of a living protocell assumes that it includes information-carrying molecules as an essential ingredient, a dividing cell-like structure can be built from a metabolism-container coupled system only. An example of such a system, modelled with dissipative particle dynamics, is presented here. This article demonstrates how a simple coupling between a precursor molecule and surfactant molecules forming micelles can experience a growth-division cycle in a predictable manner, and analyses the influence of crucial parameters on this replication cycle. Implications of these results for origins of cellular life and living technology are outlined.
H. Fellermann & R. Solé, Philos. Trans. R. Soc. Ser. B 362(1486):1803-1811, 2007
